Hi, I'm Naoto Kubota
🧬 About Me
I’m a computational biologist passionate about unraveling the complexities of life through data science and advanced computational methods. Currently, I’m working as a postdoc at Dr. Sika Zheng Lab at the University of California, Riverside.
My work bridges the gap between biology and technology, using cutting-edge computational tools to understand fundamental biological processes. Download my full CV here to learn more about my journey.
🔬 Research Focus
My research interests span multiple fascinating areas of computational biology:
- Computational genomics & transcriptomics - Analyzing large-scale genomic data to understand biological systems
- RNA splicing - Investigating how genes are processed and regulated
- Nonsense-mediated mRNA decay - Understanding quality control mechanisms in gene expression
- Neuronal development & disease in mammals - Exploring brain development and neurological disorders
- QTL mapping - Identifying genetic variants that influence traits
- Software development - Creating tools and pipelines (Bash, Python, R, Rust, Docker, Git, Snakemake, ImageJ Macro)
📚 Featured Publication
Shiba: a versatile computational method for systematic identification of differential RNA splicing across platforms
Naoto Kubota, Liang Chen, Sika Zheng Nucleic Acids Research 53(4), 2025, gkaf098
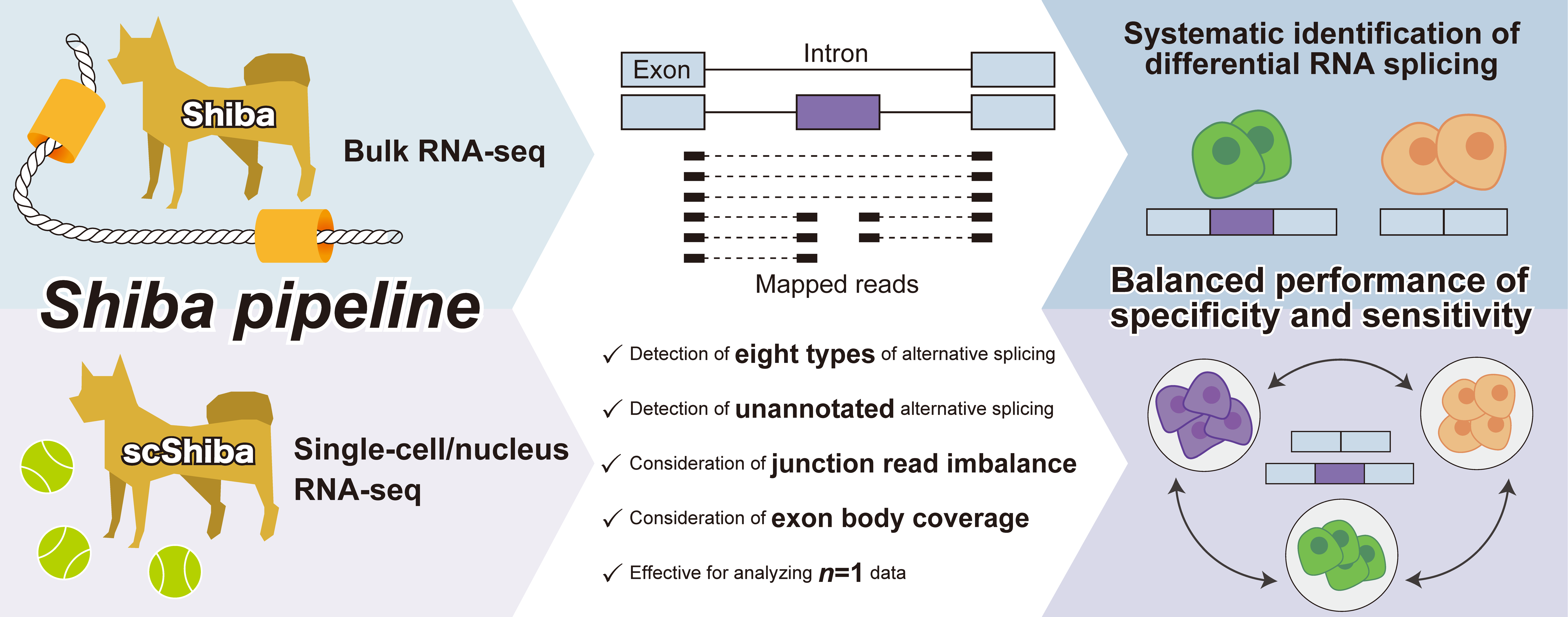
This work presents Shiba, a novel computational method that enables systematic identification of differential RNA splicing across different sequencing platforms and experimental conditions.
🎵 Beyond Science
When I’m not diving deep into genomic data, you can find me enjoying music from some incredible artists:
andymori, Quruli, Shintaro Sakamoto, Ayano Kaneko, Red Velvet, and many more amazing musicians…
Feel free to explore my research, connect with me on social media, or reach out by email!




















